Introduction
When modeling the impact of an intervention on a disease, it is common to have a first simulation phase where the intervention is disabled to achieve steady-state, followed by a second phase during which the intervention is applied. Often, we want to run the second phase many times over, varying the intervention parameters. Simulating the first phase every time is unnecessary and wasteful, since it isn’t affected by the intervention parameters.
Individual allows the user to run a simulation for a number of time steps, save the state of the simulation and resume it multiple times, with different parameters each time. This way, the initial phase before the intervention only needs to be simulated once.
Usage
The typical way to use this feature is to define a simulation
function which creates all the relevant simulation data and then calls
simulation_loop. The function we define takes in an
optional state parameter that is passed through to
simulation_loop.
run_simulation <- function(timesteps, state=NULL) {
health <- CategoricalVariable$new(c("S", "I"), rep("S", 10))
process <- bernoulli_process(health, "S", "I", 0.01)
simulation_loop(
variables=list(health),
processes=list(process),
timesteps=timesteps,
state=state)
}The simulation can be run a first time, for a given number of steps. It returns a state object, which captures the internal state of all variables and events, the state of the random number generator and the number of time steps that were simulated.
state <- run_simulation(timesteps = 50)Finally, the simulation is resumed with a larger number of time
steps, passing in the state object as an argument. The
timesteps argument refers to the total number of time
steps, including both the original simulation run and the new one. In
this case, run_simulation will only simulate 50 extra
steps. Before running the actual simulation,
simulation_loop will reload the simulation state from its
argument, overwriting any values we had set when initializing the
variables.
run_simulation(timesteps = 100, state=state)Practical example
To demonstrate the checkpoint and restore functionality of individual in a practical setting, we will use a SIRS model with a vaccination intervention. Our aim is to compare the impact of the vaccination campaign, given different vaccine efficacy scenarios.
Individuals in the simulation move from being susceptible (S) to
infectious (I) to recovered (R) and back to susceptible, after their
natural immunity wanes off. Out of the entire population N,
only I0 individuals are initially infectios, and the rest
are susceptible. Orthogonally, an individual can either be vaccinated
(Y) or not (N). The vaccination and the immunity it confers never wanes
off. All individuals are initially unvaccinated.
make_variables <- function(N, I0) {
health_states_t0 <- rep("S",N)
health_states_t0[sample.int(n = N,size = I0)] <- "I"
health <- CategoricalVariable$new(categories = c("S","I","R"), initial_values = health_states_t0)
vaccinated <- CategoricalVariable$new(categories = c("Y", "N"), initial_values = rep("N", N))
list(health=health, vaccinated=vaccinated)
}A vaccinated individual has a reduced probability of becoming infectious, as determined by the vaccine’s efficacy. The function below creates the process to model infection. It samples from the susceptible compartments, applying the different rates depending on the whether an individual’s vaccinated status.
make_infection_process <- function(health, vaccinated, N, beta, vaccine_efficacy) {
function(t) {
I <- health$get_size_of("I")
foi <- beta * I / N
vaccinated_S <- health$get_index_of("S")$and(vaccinated$get_index_of("Y"))
non_vaccinated_S <- health$get_index_of("S")$and(vaccinated$get_index_of("N"))
vaccinated_S$sample(rate = foi * (1 - vaccine_efficacy))
non_vaccinated_S$sample(rate = foi)
health$queue_update(value = "I", index = vaccinated_S)
health$queue_update(value = "I", index = non_vaccinated_S)
}
}For a while at the start of the simulation no vaccination takes
place. Only after a number of time steps, determined by the
vaccination_start parameter, does the intervention begin.
Periodically, an event will fire and every individual has a fixed
probability of becoming vaccinated. If vaccination_start is
NULL, the intervention never begins.
make_vaccination_event <- function(vaccinated, vaccination_start, vaccination_interval, vaccination_rate) {
e <- Event$new()
e$add_listener(function(t) {
print("Vaccine time")
print(t)
vaccinated$queue_update(value = "Y",
vaccinated$get_index_of("N")$sample(vaccination_rate))
e$schedule(vaccination_interval)
})
if (!is.null(vaccination_start)) {
print("Scheduling vaccination")
e$schedule(vaccination_start - 1)
}
e
}We will define our simulation as a function, taking the simulation
parameters as arguments. Any additional arguments to the function, as
denoted by ..., will be passed on to
simulation_loop. This will allow us to pass the
state argument in. The function returns the simulation data
as well as the new saved state.
run_simulation <- function(
steps,
N = 1e3,
I0 = 5,
beta = 0.1, # S -> I
gamma = 0.05, # I -> R
xi = 0.03, # R -> S
vaccination_start = NULL,
vaccination_interval = 10,
vaccination_rate = 0.05, # N -> Y
vaccine_efficacy = 1,
...)
{
variables <- make_variables(N, I0)
infection_process <- make_infection_process(
variables$health,
variables$vaccinated,
N, beta, vaccine_efficacy)
recovery_process <- bernoulli_process(variables$health, "I", "R", gamma)
return_process <- bernoulli_process(variables$health, "R", "S", xi)
vaccination_event <- make_vaccination_event(
variables$vaccinated, vaccination_start, vaccination_interval, vaccination_rate)
renderer <- Render$new(timesteps = steps)
health_render_process <- categorical_count_renderer_process(
renderer = renderer,
variable = variables$health,
categories = variables$health$get_categories()
)
processes <- list(
infection_process,
recovery_process,
return_process,
health_render_process)
final_state <- simulation_loop(
variables = variables,
events = list(vaccination_event),
processes = processes,
timesteps = steps,
...)
list(result=renderer$to_dataframe(), state=final_state)
}We will start by running and plotting our baseline simulation, with the intervention disabled.
data <- run_simulation(steps=1500)$result
colours <- c("royalblue3","firebrick3","darkorchid3")
matplot(
x=data["timestep"],
y=data[c("S_count","I_count", "R_count")],
xlab="Time", ylab="Count",
type="l", lwd=2, lty = 1, col = colours
)
legend(
x = "topright",
pch = rep(16,3),
col = colours,
legend = c("S", "I", "R"), cex = 1.5,
bg='white'
)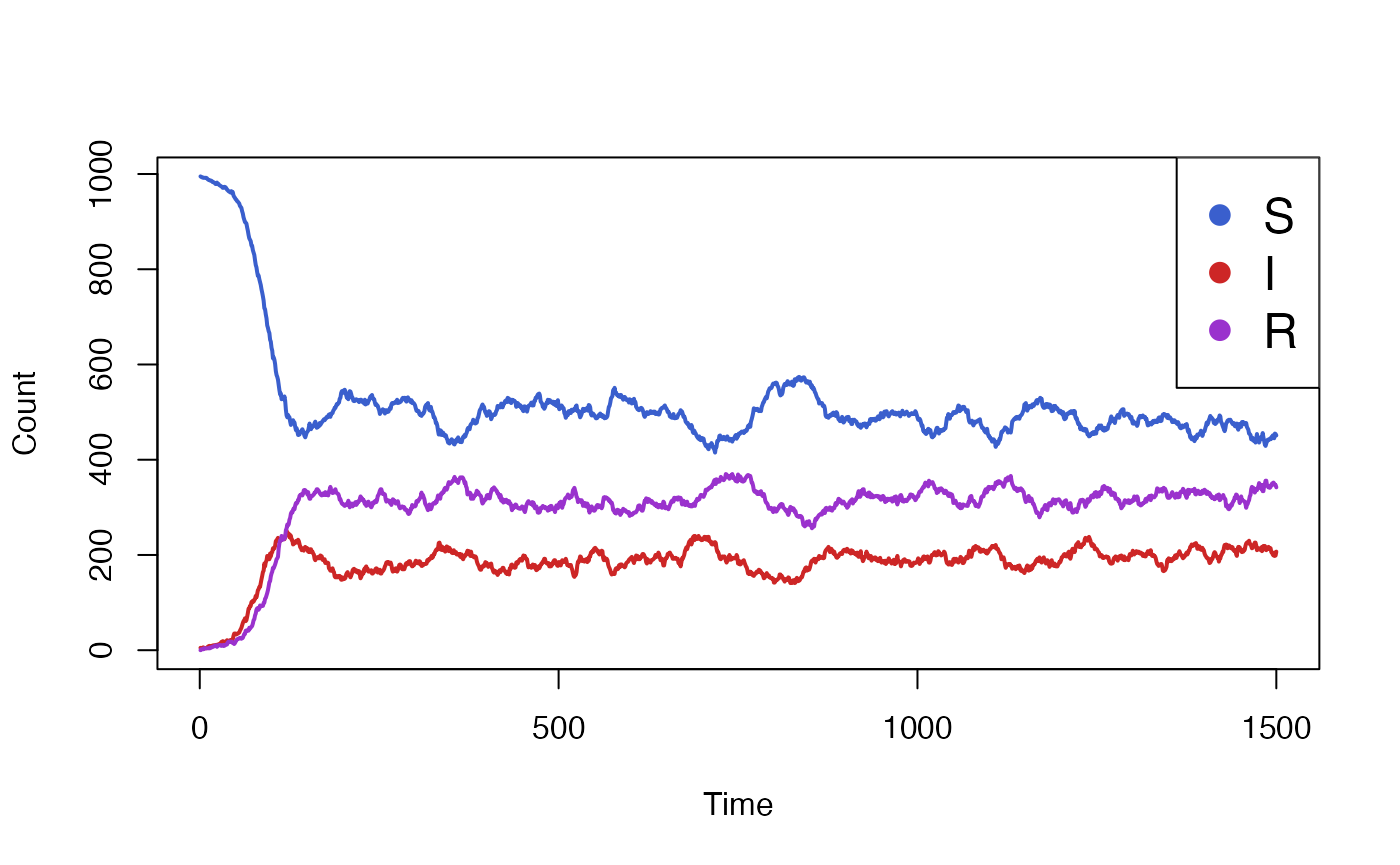
We see that the simulation takes some time to settle from its initial
parameters to its steady-state conditions. We will now enable the
vaccine intervention, but only starting at a point after the simulation
has settled, for example at t=500.
data <- run_simulation(steps=1500, vaccination_start = 500, vaccine_efficacy = 1)$result
#> [1] "Scheduling vaccination"
#> [1] "Vaccine time"
#> [1] 500
#> [1] "Vaccine time"
#> [1] 510
#> [1] "Vaccine time"
#> [1] 520
#> [1] "Vaccine time"
#> [1] 530
#> [1] "Vaccine time"
#> [1] 540
#> [1] "Vaccine time"
#> [1] 550
#> [1] "Vaccine time"
#> [1] 560
#> [1] "Vaccine time"
#> [1] 570
#> [1] "Vaccine time"
#> [1] 580
#> [1] "Vaccine time"
#> [1] 590
#> [1] "Vaccine time"
#> [1] 600
#> [1] "Vaccine time"
#> [1] 610
#> [1] "Vaccine time"
#> [1] 620
#> [1] "Vaccine time"
#> [1] 630
#> [1] "Vaccine time"
#> [1] 640
#> [1] "Vaccine time"
#> [1] 650
#> [1] "Vaccine time"
#> [1] 660
#> [1] "Vaccine time"
#> [1] 670
#> [1] "Vaccine time"
#> [1] 680
#> [1] "Vaccine time"
#> [1] 690
#> [1] "Vaccine time"
#> [1] 700
#> [1] "Vaccine time"
#> [1] 710
#> [1] "Vaccine time"
#> [1] 720
#> [1] "Vaccine time"
#> [1] 730
#> [1] "Vaccine time"
#> [1] 740
#> [1] "Vaccine time"
#> [1] 750
#> [1] "Vaccine time"
#> [1] 760
#> [1] "Vaccine time"
#> [1] 770
#> [1] "Vaccine time"
#> [1] 780
#> [1] "Vaccine time"
#> [1] 790
#> [1] "Vaccine time"
#> [1] 800
#> [1] "Vaccine time"
#> [1] 810
#> [1] "Vaccine time"
#> [1] 820
#> [1] "Vaccine time"
#> [1] 830
#> [1] "Vaccine time"
#> [1] 840
#> [1] "Vaccine time"
#> [1] 850
#> [1] "Vaccine time"
#> [1] 860
#> [1] "Vaccine time"
#> [1] 870
#> [1] "Vaccine time"
#> [1] 880
#> [1] "Vaccine time"
#> [1] 890
#> [1] "Vaccine time"
#> [1] 900
#> [1] "Vaccine time"
#> [1] 910
#> [1] "Vaccine time"
#> [1] 920
#> [1] "Vaccine time"
#> [1] 930
#> [1] "Vaccine time"
#> [1] 940
#> [1] "Vaccine time"
#> [1] 950
#> [1] "Vaccine time"
#> [1] 960
#> [1] "Vaccine time"
#> [1] 970
#> [1] "Vaccine time"
#> [1] 980
#> [1] "Vaccine time"
#> [1] 990
#> [1] "Vaccine time"
#> [1] 1000
#> [1] "Vaccine time"
#> [1] 1010
#> [1] "Vaccine time"
#> [1] 1020
#> [1] "Vaccine time"
#> [1] 1030
#> [1] "Vaccine time"
#> [1] 1040
#> [1] "Vaccine time"
#> [1] 1050
#> [1] "Vaccine time"
#> [1] 1060
#> [1] "Vaccine time"
#> [1] 1070
#> [1] "Vaccine time"
#> [1] 1080
#> [1] "Vaccine time"
#> [1] 1090
#> [1] "Vaccine time"
#> [1] 1100
#> [1] "Vaccine time"
#> [1] 1110
#> [1] "Vaccine time"
#> [1] 1120
#> [1] "Vaccine time"
#> [1] 1130
#> [1] "Vaccine time"
#> [1] 1140
#> [1] "Vaccine time"
#> [1] 1150
#> [1] "Vaccine time"
#> [1] 1160
#> [1] "Vaccine time"
#> [1] 1170
#> [1] "Vaccine time"
#> [1] 1180
#> [1] "Vaccine time"
#> [1] 1190
#> [1] "Vaccine time"
#> [1] 1200
#> [1] "Vaccine time"
#> [1] 1210
#> [1] "Vaccine time"
#> [1] 1220
#> [1] "Vaccine time"
#> [1] 1230
#> [1] "Vaccine time"
#> [1] 1240
#> [1] "Vaccine time"
#> [1] 1250
#> [1] "Vaccine time"
#> [1] 1260
#> [1] "Vaccine time"
#> [1] 1270
#> [1] "Vaccine time"
#> [1] 1280
#> [1] "Vaccine time"
#> [1] 1290
#> [1] "Vaccine time"
#> [1] 1300
#> [1] "Vaccine time"
#> [1] 1310
#> [1] "Vaccine time"
#> [1] 1320
#> [1] "Vaccine time"
#> [1] 1330
#> [1] "Vaccine time"
#> [1] 1340
#> [1] "Vaccine time"
#> [1] 1350
#> [1] "Vaccine time"
#> [1] 1360
#> [1] "Vaccine time"
#> [1] 1370
#> [1] "Vaccine time"
#> [1] 1380
#> [1] "Vaccine time"
#> [1] 1390
#> [1] "Vaccine time"
#> [1] 1400
#> [1] "Vaccine time"
#> [1] 1410
#> [1] "Vaccine time"
#> [1] 1420
#> [1] "Vaccine time"
#> [1] 1430
#> [1] "Vaccine time"
#> [1] 1440
#> [1] "Vaccine time"
#> [1] 1450
#> [1] "Vaccine time"
#> [1] 1460
#> [1] "Vaccine time"
#> [1] 1470
#> [1] "Vaccine time"
#> [1] 1480
#> [1] "Vaccine time"
#> [1] 1490
#> [1] "Vaccine time"
#> [1] 1500
colours <- c("royalblue3","firebrick3","darkorchid3")
matplot(
x=data["timestep"],
y=data[c("S_count","I_count", "R_count")],
xlab="Time", ylab="Count",
type="l", lwd=2, lty = 1, col = colours
)
legend(
x = "topright",
pch = rep(16,3),
col = colours,
legend = c("S", "I", "R"), cex = 1.5,
bg='white'
)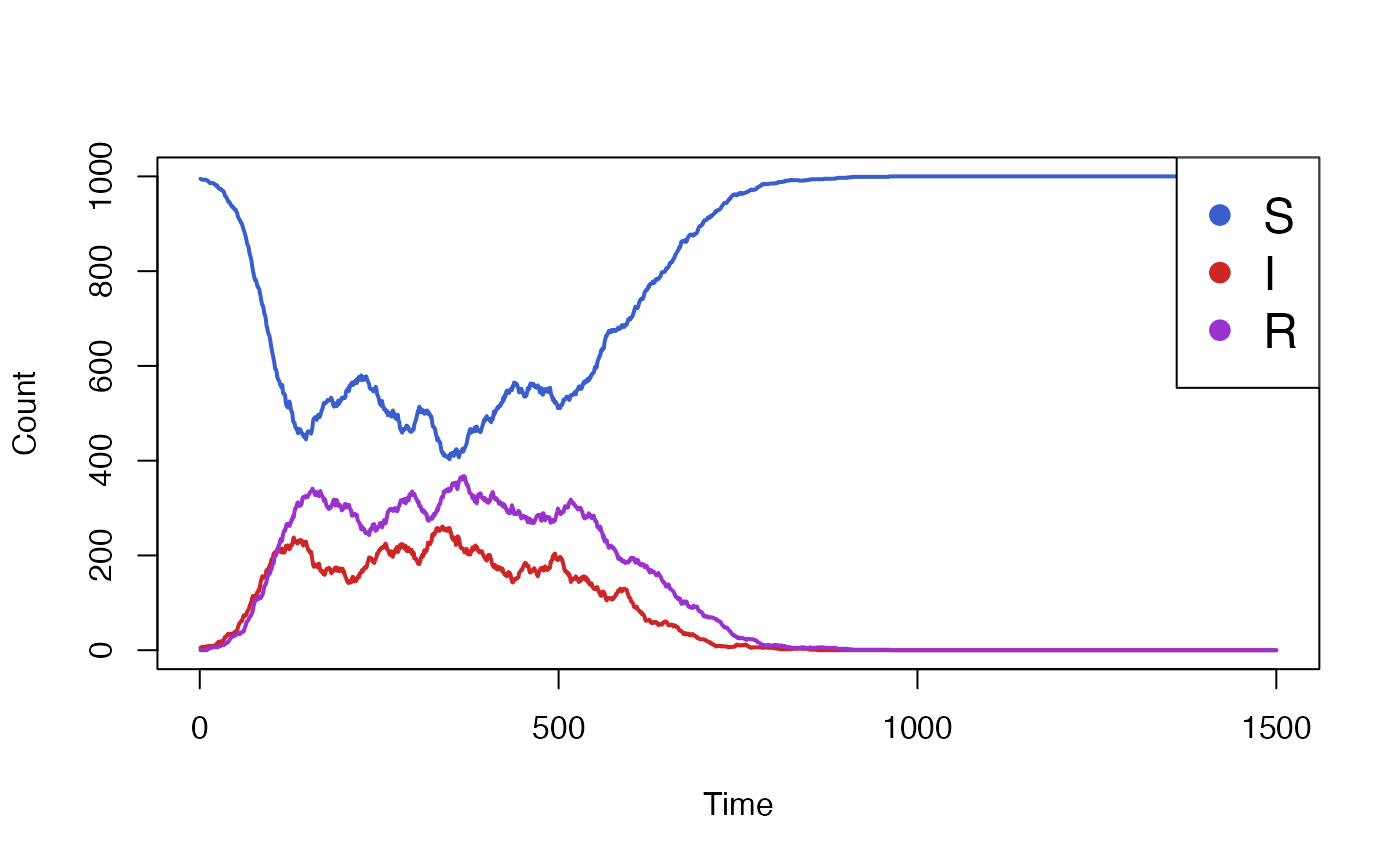
The simulation above clearly shows the effect of the vaccination
campaign, starting at t=500. However, it made the
optimistic assumption of a 100% vaccine efficacy. We wish to run the
simulation again but with varying levels of efficacy, in order the
compare its impact.
While we could run the code above many times over, each simulation
would repeat the first 499 timesteps, despite the result being identical
each time. Instead we start by running only these timesteps, and saving
the result. We do need to specify the start of the intervention, as it
is necessary to schedule the first vaccination event. However the
details of the intervention (ie. vaccine_efficacy) are
irrelevant and can be omitted.
initial <- run_simulation(steps=499, vaccination_start = 500)
#> [1] "Scheduling vaccination"From this initial result, we can resume the simulation, but using different values of vaccine efficacy each time. We also include a control simulation, in which no vaccination takes place. Each of these simulation will skip the first 499 steps and only run the next 1001 time steps.
control <- run_simulation(steps=1500, vaccination_start = 500, vaccine_efficacy=0.0, state=initial$state)
#> [1] "Scheduling vaccination"
#> [1] "Vaccine time"
#> [1] 500
#> [1] "Vaccine time"
#> [1] 510
#> [1] "Vaccine time"
#> [1] 520
#> [1] "Vaccine time"
#> [1] 530
#> [1] "Vaccine time"
#> [1] 540
#> [1] "Vaccine time"
#> [1] 550
#> [1] "Vaccine time"
#> [1] 560
#> [1] "Vaccine time"
#> [1] 570
#> [1] "Vaccine time"
#> [1] 580
#> [1] "Vaccine time"
#> [1] 590
#> [1] "Vaccine time"
#> [1] 600
#> [1] "Vaccine time"
#> [1] 610
#> [1] "Vaccine time"
#> [1] 620
#> [1] "Vaccine time"
#> [1] 630
#> [1] "Vaccine time"
#> [1] 640
#> [1] "Vaccine time"
#> [1] 650
#> [1] "Vaccine time"
#> [1] 660
#> [1] "Vaccine time"
#> [1] 670
#> [1] "Vaccine time"
#> [1] 680
#> [1] "Vaccine time"
#> [1] 690
#> [1] "Vaccine time"
#> [1] 700
#> [1] "Vaccine time"
#> [1] 710
#> [1] "Vaccine time"
#> [1] 720
#> [1] "Vaccine time"
#> [1] 730
#> [1] "Vaccine time"
#> [1] 740
#> [1] "Vaccine time"
#> [1] 750
#> [1] "Vaccine time"
#> [1] 760
#> [1] "Vaccine time"
#> [1] 770
#> [1] "Vaccine time"
#> [1] 780
#> [1] "Vaccine time"
#> [1] 790
#> [1] "Vaccine time"
#> [1] 800
#> [1] "Vaccine time"
#> [1] 810
#> [1] "Vaccine time"
#> [1] 820
#> [1] "Vaccine time"
#> [1] 830
#> [1] "Vaccine time"
#> [1] 840
#> [1] "Vaccine time"
#> [1] 850
#> [1] "Vaccine time"
#> [1] 860
#> [1] "Vaccine time"
#> [1] 870
#> [1] "Vaccine time"
#> [1] 880
#> [1] "Vaccine time"
#> [1] 890
#> [1] "Vaccine time"
#> [1] 900
#> [1] "Vaccine time"
#> [1] 910
#> [1] "Vaccine time"
#> [1] 920
#> [1] "Vaccine time"
#> [1] 930
#> [1] "Vaccine time"
#> [1] 940
#> [1] "Vaccine time"
#> [1] 950
#> [1] "Vaccine time"
#> [1] 960
#> [1] "Vaccine time"
#> [1] 970
#> [1] "Vaccine time"
#> [1] 980
#> [1] "Vaccine time"
#> [1] 990
#> [1] "Vaccine time"
#> [1] 1000
#> [1] "Vaccine time"
#> [1] 1010
#> [1] "Vaccine time"
#> [1] 1020
#> [1] "Vaccine time"
#> [1] 1030
#> [1] "Vaccine time"
#> [1] 1040
#> [1] "Vaccine time"
#> [1] 1050
#> [1] "Vaccine time"
#> [1] 1060
#> [1] "Vaccine time"
#> [1] 1070
#> [1] "Vaccine time"
#> [1] 1080
#> [1] "Vaccine time"
#> [1] 1090
#> [1] "Vaccine time"
#> [1] 1100
#> [1] "Vaccine time"
#> [1] 1110
#> [1] "Vaccine time"
#> [1] 1120
#> [1] "Vaccine time"
#> [1] 1130
#> [1] "Vaccine time"
#> [1] 1140
#> [1] "Vaccine time"
#> [1] 1150
#> [1] "Vaccine time"
#> [1] 1160
#> [1] "Vaccine time"
#> [1] 1170
#> [1] "Vaccine time"
#> [1] 1180
#> [1] "Vaccine time"
#> [1] 1190
#> [1] "Vaccine time"
#> [1] 1200
#> [1] "Vaccine time"
#> [1] 1210
#> [1] "Vaccine time"
#> [1] 1220
#> [1] "Vaccine time"
#> [1] 1230
#> [1] "Vaccine time"
#> [1] 1240
#> [1] "Vaccine time"
#> [1] 1250
#> [1] "Vaccine time"
#> [1] 1260
#> [1] "Vaccine time"
#> [1] 1270
#> [1] "Vaccine time"
#> [1] 1280
#> [1] "Vaccine time"
#> [1] 1290
#> [1] "Vaccine time"
#> [1] 1300
#> [1] "Vaccine time"
#> [1] 1310
#> [1] "Vaccine time"
#> [1] 1320
#> [1] "Vaccine time"
#> [1] 1330
#> [1] "Vaccine time"
#> [1] 1340
#> [1] "Vaccine time"
#> [1] 1350
#> [1] "Vaccine time"
#> [1] 1360
#> [1] "Vaccine time"
#> [1] 1370
#> [1] "Vaccine time"
#> [1] 1380
#> [1] "Vaccine time"
#> [1] 1390
#> [1] "Vaccine time"
#> [1] 1400
#> [1] "Vaccine time"
#> [1] 1410
#> [1] "Vaccine time"
#> [1] 1420
#> [1] "Vaccine time"
#> [1] 1430
#> [1] "Vaccine time"
#> [1] 1440
#> [1] "Vaccine time"
#> [1] 1450
#> [1] "Vaccine time"
#> [1] 1460
#> [1] "Vaccine time"
#> [1] 1470
#> [1] "Vaccine time"
#> [1] 1480
#> [1] "Vaccine time"
#> [1] 1490
#> [1] "Vaccine time"
#> [1] 1500
vaccine30 <- run_simulation(steps=1500, vaccination_start = 500, vaccine_efficacy=0.3, state=initial$state)
#> [1] "Scheduling vaccination"
#> [1] "Vaccine time"
#> [1] 500
#> [1] "Vaccine time"
#> [1] 510
#> [1] "Vaccine time"
#> [1] 520
#> [1] "Vaccine time"
#> [1] 530
#> [1] "Vaccine time"
#> [1] 540
#> [1] "Vaccine time"
#> [1] 550
#> [1] "Vaccine time"
#> [1] 560
#> [1] "Vaccine time"
#> [1] 570
#> [1] "Vaccine time"
#> [1] 580
#> [1] "Vaccine time"
#> [1] 590
#> [1] "Vaccine time"
#> [1] 600
#> [1] "Vaccine time"
#> [1] 610
#> [1] "Vaccine time"
#> [1] 620
#> [1] "Vaccine time"
#> [1] 630
#> [1] "Vaccine time"
#> [1] 640
#> [1] "Vaccine time"
#> [1] 650
#> [1] "Vaccine time"
#> [1] 660
#> [1] "Vaccine time"
#> [1] 670
#> [1] "Vaccine time"
#> [1] 680
#> [1] "Vaccine time"
#> [1] 690
#> [1] "Vaccine time"
#> [1] 700
#> [1] "Vaccine time"
#> [1] 710
#> [1] "Vaccine time"
#> [1] 720
#> [1] "Vaccine time"
#> [1] 730
#> [1] "Vaccine time"
#> [1] 740
#> [1] "Vaccine time"
#> [1] 750
#> [1] "Vaccine time"
#> [1] 760
#> [1] "Vaccine time"
#> [1] 770
#> [1] "Vaccine time"
#> [1] 780
#> [1] "Vaccine time"
#> [1] 790
#> [1] "Vaccine time"
#> [1] 800
#> [1] "Vaccine time"
#> [1] 810
#> [1] "Vaccine time"
#> [1] 820
#> [1] "Vaccine time"
#> [1] 830
#> [1] "Vaccine time"
#> [1] 840
#> [1] "Vaccine time"
#> [1] 850
#> [1] "Vaccine time"
#> [1] 860
#> [1] "Vaccine time"
#> [1] 870
#> [1] "Vaccine time"
#> [1] 880
#> [1] "Vaccine time"
#> [1] 890
#> [1] "Vaccine time"
#> [1] 900
#> [1] "Vaccine time"
#> [1] 910
#> [1] "Vaccine time"
#> [1] 920
#> [1] "Vaccine time"
#> [1] 930
#> [1] "Vaccine time"
#> [1] 940
#> [1] "Vaccine time"
#> [1] 950
#> [1] "Vaccine time"
#> [1] 960
#> [1] "Vaccine time"
#> [1] 970
#> [1] "Vaccine time"
#> [1] 980
#> [1] "Vaccine time"
#> [1] 990
#> [1] "Vaccine time"
#> [1] 1000
#> [1] "Vaccine time"
#> [1] 1010
#> [1] "Vaccine time"
#> [1] 1020
#> [1] "Vaccine time"
#> [1] 1030
#> [1] "Vaccine time"
#> [1] 1040
#> [1] "Vaccine time"
#> [1] 1050
#> [1] "Vaccine time"
#> [1] 1060
#> [1] "Vaccine time"
#> [1] 1070
#> [1] "Vaccine time"
#> [1] 1080
#> [1] "Vaccine time"
#> [1] 1090
#> [1] "Vaccine time"
#> [1] 1100
#> [1] "Vaccine time"
#> [1] 1110
#> [1] "Vaccine time"
#> [1] 1120
#> [1] "Vaccine time"
#> [1] 1130
#> [1] "Vaccine time"
#> [1] 1140
#> [1] "Vaccine time"
#> [1] 1150
#> [1] "Vaccine time"
#> [1] 1160
#> [1] "Vaccine time"
#> [1] 1170
#> [1] "Vaccine time"
#> [1] 1180
#> [1] "Vaccine time"
#> [1] 1190
#> [1] "Vaccine time"
#> [1] 1200
#> [1] "Vaccine time"
#> [1] 1210
#> [1] "Vaccine time"
#> [1] 1220
#> [1] "Vaccine time"
#> [1] 1230
#> [1] "Vaccine time"
#> [1] 1240
#> [1] "Vaccine time"
#> [1] 1250
#> [1] "Vaccine time"
#> [1] 1260
#> [1] "Vaccine time"
#> [1] 1270
#> [1] "Vaccine time"
#> [1] 1280
#> [1] "Vaccine time"
#> [1] 1290
#> [1] "Vaccine time"
#> [1] 1300
#> [1] "Vaccine time"
#> [1] 1310
#> [1] "Vaccine time"
#> [1] 1320
#> [1] "Vaccine time"
#> [1] 1330
#> [1] "Vaccine time"
#> [1] 1340
#> [1] "Vaccine time"
#> [1] 1350
#> [1] "Vaccine time"
#> [1] 1360
#> [1] "Vaccine time"
#> [1] 1370
#> [1] "Vaccine time"
#> [1] 1380
#> [1] "Vaccine time"
#> [1] 1390
#> [1] "Vaccine time"
#> [1] 1400
#> [1] "Vaccine time"
#> [1] 1410
#> [1] "Vaccine time"
#> [1] 1420
#> [1] "Vaccine time"
#> [1] 1430
#> [1] "Vaccine time"
#> [1] 1440
#> [1] "Vaccine time"
#> [1] 1450
#> [1] "Vaccine time"
#> [1] 1460
#> [1] "Vaccine time"
#> [1] 1470
#> [1] "Vaccine time"
#> [1] 1480
#> [1] "Vaccine time"
#> [1] 1490
#> [1] "Vaccine time"
#> [1] 1500
vaccine50 <- run_simulation(steps=1500, vaccination_start = 500, vaccine_efficacy=0.5, state=initial$state)
#> [1] "Scheduling vaccination"
#> [1] "Vaccine time"
#> [1] 500
#> [1] "Vaccine time"
#> [1] 510
#> [1] "Vaccine time"
#> [1] 520
#> [1] "Vaccine time"
#> [1] 530
#> [1] "Vaccine time"
#> [1] 540
#> [1] "Vaccine time"
#> [1] 550
#> [1] "Vaccine time"
#> [1] 560
#> [1] "Vaccine time"
#> [1] 570
#> [1] "Vaccine time"
#> [1] 580
#> [1] "Vaccine time"
#> [1] 590
#> [1] "Vaccine time"
#> [1] 600
#> [1] "Vaccine time"
#> [1] 610
#> [1] "Vaccine time"
#> [1] 620
#> [1] "Vaccine time"
#> [1] 630
#> [1] "Vaccine time"
#> [1] 640
#> [1] "Vaccine time"
#> [1] 650
#> [1] "Vaccine time"
#> [1] 660
#> [1] "Vaccine time"
#> [1] 670
#> [1] "Vaccine time"
#> [1] 680
#> [1] "Vaccine time"
#> [1] 690
#> [1] "Vaccine time"
#> [1] 700
#> [1] "Vaccine time"
#> [1] 710
#> [1] "Vaccine time"
#> [1] 720
#> [1] "Vaccine time"
#> [1] 730
#> [1] "Vaccine time"
#> [1] 740
#> [1] "Vaccine time"
#> [1] 750
#> [1] "Vaccine time"
#> [1] 760
#> [1] "Vaccine time"
#> [1] 770
#> [1] "Vaccine time"
#> [1] 780
#> [1] "Vaccine time"
#> [1] 790
#> [1] "Vaccine time"
#> [1] 800
#> [1] "Vaccine time"
#> [1] 810
#> [1] "Vaccine time"
#> [1] 820
#> [1] "Vaccine time"
#> [1] 830
#> [1] "Vaccine time"
#> [1] 840
#> [1] "Vaccine time"
#> [1] 850
#> [1] "Vaccine time"
#> [1] 860
#> [1] "Vaccine time"
#> [1] 870
#> [1] "Vaccine time"
#> [1] 880
#> [1] "Vaccine time"
#> [1] 890
#> [1] "Vaccine time"
#> [1] 900
#> [1] "Vaccine time"
#> [1] 910
#> [1] "Vaccine time"
#> [1] 920
#> [1] "Vaccine time"
#> [1] 930
#> [1] "Vaccine time"
#> [1] 940
#> [1] "Vaccine time"
#> [1] 950
#> [1] "Vaccine time"
#> [1] 960
#> [1] "Vaccine time"
#> [1] 970
#> [1] "Vaccine time"
#> [1] 980
#> [1] "Vaccine time"
#> [1] 990
#> [1] "Vaccine time"
#> [1] 1000
#> [1] "Vaccine time"
#> [1] 1010
#> [1] "Vaccine time"
#> [1] 1020
#> [1] "Vaccine time"
#> [1] 1030
#> [1] "Vaccine time"
#> [1] 1040
#> [1] "Vaccine time"
#> [1] 1050
#> [1] "Vaccine time"
#> [1] 1060
#> [1] "Vaccine time"
#> [1] 1070
#> [1] "Vaccine time"
#> [1] 1080
#> [1] "Vaccine time"
#> [1] 1090
#> [1] "Vaccine time"
#> [1] 1100
#> [1] "Vaccine time"
#> [1] 1110
#> [1] "Vaccine time"
#> [1] 1120
#> [1] "Vaccine time"
#> [1] 1130
#> [1] "Vaccine time"
#> [1] 1140
#> [1] "Vaccine time"
#> [1] 1150
#> [1] "Vaccine time"
#> [1] 1160
#> [1] "Vaccine time"
#> [1] 1170
#> [1] "Vaccine time"
#> [1] 1180
#> [1] "Vaccine time"
#> [1] 1190
#> [1] "Vaccine time"
#> [1] 1200
#> [1] "Vaccine time"
#> [1] 1210
#> [1] "Vaccine time"
#> [1] 1220
#> [1] "Vaccine time"
#> [1] 1230
#> [1] "Vaccine time"
#> [1] 1240
#> [1] "Vaccine time"
#> [1] 1250
#> [1] "Vaccine time"
#> [1] 1260
#> [1] "Vaccine time"
#> [1] 1270
#> [1] "Vaccine time"
#> [1] 1280
#> [1] "Vaccine time"
#> [1] 1290
#> [1] "Vaccine time"
#> [1] 1300
#> [1] "Vaccine time"
#> [1] 1310
#> [1] "Vaccine time"
#> [1] 1320
#> [1] "Vaccine time"
#> [1] 1330
#> [1] "Vaccine time"
#> [1] 1340
#> [1] "Vaccine time"
#> [1] 1350
#> [1] "Vaccine time"
#> [1] 1360
#> [1] "Vaccine time"
#> [1] 1370
#> [1] "Vaccine time"
#> [1] 1380
#> [1] "Vaccine time"
#> [1] 1390
#> [1] "Vaccine time"
#> [1] 1400
#> [1] "Vaccine time"
#> [1] 1410
#> [1] "Vaccine time"
#> [1] 1420
#> [1] "Vaccine time"
#> [1] 1430
#> [1] "Vaccine time"
#> [1] 1440
#> [1] "Vaccine time"
#> [1] 1450
#> [1] "Vaccine time"
#> [1] 1460
#> [1] "Vaccine time"
#> [1] 1470
#> [1] "Vaccine time"
#> [1] 1480
#> [1] "Vaccine time"
#> [1] 1490
#> [1] "Vaccine time"
#> [1] 1500
vaccine100 <- run_simulation(steps=1500, vaccination_start = 500, vaccine_efficacy=1.0, state=initial$state)
#> [1] "Scheduling vaccination"
#> [1] "Vaccine time"
#> [1] 500
#> [1] "Vaccine time"
#> [1] 510
#> [1] "Vaccine time"
#> [1] 520
#> [1] "Vaccine time"
#> [1] 530
#> [1] "Vaccine time"
#> [1] 540
#> [1] "Vaccine time"
#> [1] 550
#> [1] "Vaccine time"
#> [1] 560
#> [1] "Vaccine time"
#> [1] 570
#> [1] "Vaccine time"
#> [1] 580
#> [1] "Vaccine time"
#> [1] 590
#> [1] "Vaccine time"
#> [1] 600
#> [1] "Vaccine time"
#> [1] 610
#> [1] "Vaccine time"
#> [1] 620
#> [1] "Vaccine time"
#> [1] 630
#> [1] "Vaccine time"
#> [1] 640
#> [1] "Vaccine time"
#> [1] 650
#> [1] "Vaccine time"
#> [1] 660
#> [1] "Vaccine time"
#> [1] 670
#> [1] "Vaccine time"
#> [1] 680
#> [1] "Vaccine time"
#> [1] 690
#> [1] "Vaccine time"
#> [1] 700
#> [1] "Vaccine time"
#> [1] 710
#> [1] "Vaccine time"
#> [1] 720
#> [1] "Vaccine time"
#> [1] 730
#> [1] "Vaccine time"
#> [1] 740
#> [1] "Vaccine time"
#> [1] 750
#> [1] "Vaccine time"
#> [1] 760
#> [1] "Vaccine time"
#> [1] 770
#> [1] "Vaccine time"
#> [1] 780
#> [1] "Vaccine time"
#> [1] 790
#> [1] "Vaccine time"
#> [1] 800
#> [1] "Vaccine time"
#> [1] 810
#> [1] "Vaccine time"
#> [1] 820
#> [1] "Vaccine time"
#> [1] 830
#> [1] "Vaccine time"
#> [1] 840
#> [1] "Vaccine time"
#> [1] 850
#> [1] "Vaccine time"
#> [1] 860
#> [1] "Vaccine time"
#> [1] 870
#> [1] "Vaccine time"
#> [1] 880
#> [1] "Vaccine time"
#> [1] 890
#> [1] "Vaccine time"
#> [1] 900
#> [1] "Vaccine time"
#> [1] 910
#> [1] "Vaccine time"
#> [1] 920
#> [1] "Vaccine time"
#> [1] 930
#> [1] "Vaccine time"
#> [1] 940
#> [1] "Vaccine time"
#> [1] 950
#> [1] "Vaccine time"
#> [1] 960
#> [1] "Vaccine time"
#> [1] 970
#> [1] "Vaccine time"
#> [1] 980
#> [1] "Vaccine time"
#> [1] 990
#> [1] "Vaccine time"
#> [1] 1000
#> [1] "Vaccine time"
#> [1] 1010
#> [1] "Vaccine time"
#> [1] 1020
#> [1] "Vaccine time"
#> [1] 1030
#> [1] "Vaccine time"
#> [1] 1040
#> [1] "Vaccine time"
#> [1] 1050
#> [1] "Vaccine time"
#> [1] 1060
#> [1] "Vaccine time"
#> [1] 1070
#> [1] "Vaccine time"
#> [1] 1080
#> [1] "Vaccine time"
#> [1] 1090
#> [1] "Vaccine time"
#> [1] 1100
#> [1] "Vaccine time"
#> [1] 1110
#> [1] "Vaccine time"
#> [1] 1120
#> [1] "Vaccine time"
#> [1] 1130
#> [1] "Vaccine time"
#> [1] 1140
#> [1] "Vaccine time"
#> [1] 1150
#> [1] "Vaccine time"
#> [1] 1160
#> [1] "Vaccine time"
#> [1] 1170
#> [1] "Vaccine time"
#> [1] 1180
#> [1] "Vaccine time"
#> [1] 1190
#> [1] "Vaccine time"
#> [1] 1200
#> [1] "Vaccine time"
#> [1] 1210
#> [1] "Vaccine time"
#> [1] 1220
#> [1] "Vaccine time"
#> [1] 1230
#> [1] "Vaccine time"
#> [1] 1240
#> [1] "Vaccine time"
#> [1] 1250
#> [1] "Vaccine time"
#> [1] 1260
#> [1] "Vaccine time"
#> [1] 1270
#> [1] "Vaccine time"
#> [1] 1280
#> [1] "Vaccine time"
#> [1] 1290
#> [1] "Vaccine time"
#> [1] 1300
#> [1] "Vaccine time"
#> [1] 1310
#> [1] "Vaccine time"
#> [1] 1320
#> [1] "Vaccine time"
#> [1] 1330
#> [1] "Vaccine time"
#> [1] 1340
#> [1] "Vaccine time"
#> [1] 1350
#> [1] "Vaccine time"
#> [1] 1360
#> [1] "Vaccine time"
#> [1] 1370
#> [1] "Vaccine time"
#> [1] 1380
#> [1] "Vaccine time"
#> [1] 1390
#> [1] "Vaccine time"
#> [1] 1400
#> [1] "Vaccine time"
#> [1] 1410
#> [1] "Vaccine time"
#> [1] 1420
#> [1] "Vaccine time"
#> [1] 1430
#> [1] "Vaccine time"
#> [1] 1440
#> [1] "Vaccine time"
#> [1] 1450
#> [1] "Vaccine time"
#> [1] 1460
#> [1] "Vaccine time"
#> [1] 1470
#> [1] "Vaccine time"
#> [1] 1480
#> [1] "Vaccine time"
#> [1] 1490
#> [1] "Vaccine time"
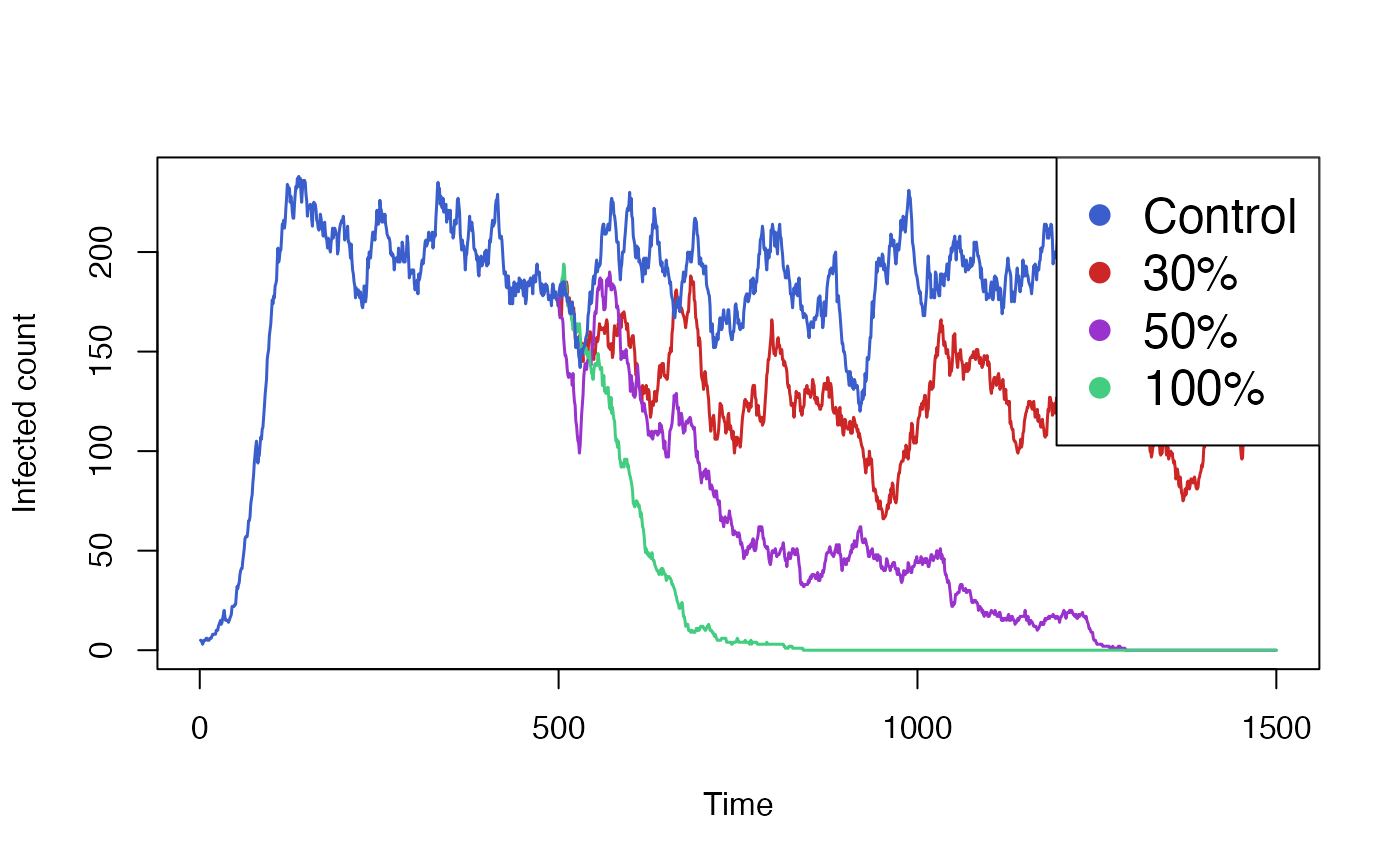
#> [1] 1500Finally we aggregate and plot the results from all these simulations. We also need to include the data from our initial run, which we will plot the same colour as our control simulation.
colours <- c("royalblue3","firebrick3","darkorchid3", "seagreen3")
# Pad initial out to ensure it has the same shape as other series.
initial$result[500:1500,] <- NA
matplot(
data.frame(
initial$result[,"I_count"],
vaccine30$result[,"I_count"],
vaccine50$result[,"I_count"],
vaccine100$result[,"I_count"],
control$result[,"I_count"]),
xlab = "Time", ylab = "Infected count",
type = "l", lwd = 1.5, lty = 1, col = colours,
)
legend(
x = "topright", pch = rep(16,3),
col = colours,
legend = c("Control", "30%", "50%", "100%"), cex = 1.5,
bg='white'
)
Caveats
Saving and restoring the simulation state comes with a number of caveats.
- All simulation state must be represented as objects managed by individual. Any state maintained externally will not be saved nor restored.
- The state object’s structure is not stable and is expected to change. One should not expect to serialize the state to disk and have it work with future versions of the individual package.
- The simulation must be re-created in an identical way. Variables and
events may not be added or removed, variable sizes must remain constant,
the list of categories in a
CategoricalVariablecannot be modified, etc. The order of variables and events passed to therun_simulationfunction must remain stable. - If an event is scheduled before the checkpoint, the time at which it
will execute cannot be changed when resuming, even if that time is in
the future. For example in the SIRS model above, we would not be able to
resume the simulation with different values for
vaccination_start; changing that parameter would have no effect.
While parameters of the simulation can be changed between the initial
run and the subsequent runs (as demonstrated with the
vaccine_efficacy parameter above), in general you should
not modify parameters that would have been already had an impact on the
first part of the simulation. Doing so would produce results that can
only be produced through checkpoint and resume, and not as a single
simulation.
For example, in our SIRS model, it may be tempting to model a time-varying parameter by running half of the simulation with one value and then resuming it with a different value. While this would probably work, it would be brittle and hard to compose. As more time-varying parameters are introduced to the model, the simulation would need to be saved and restored each time a value changes.
Restoring random number generator state
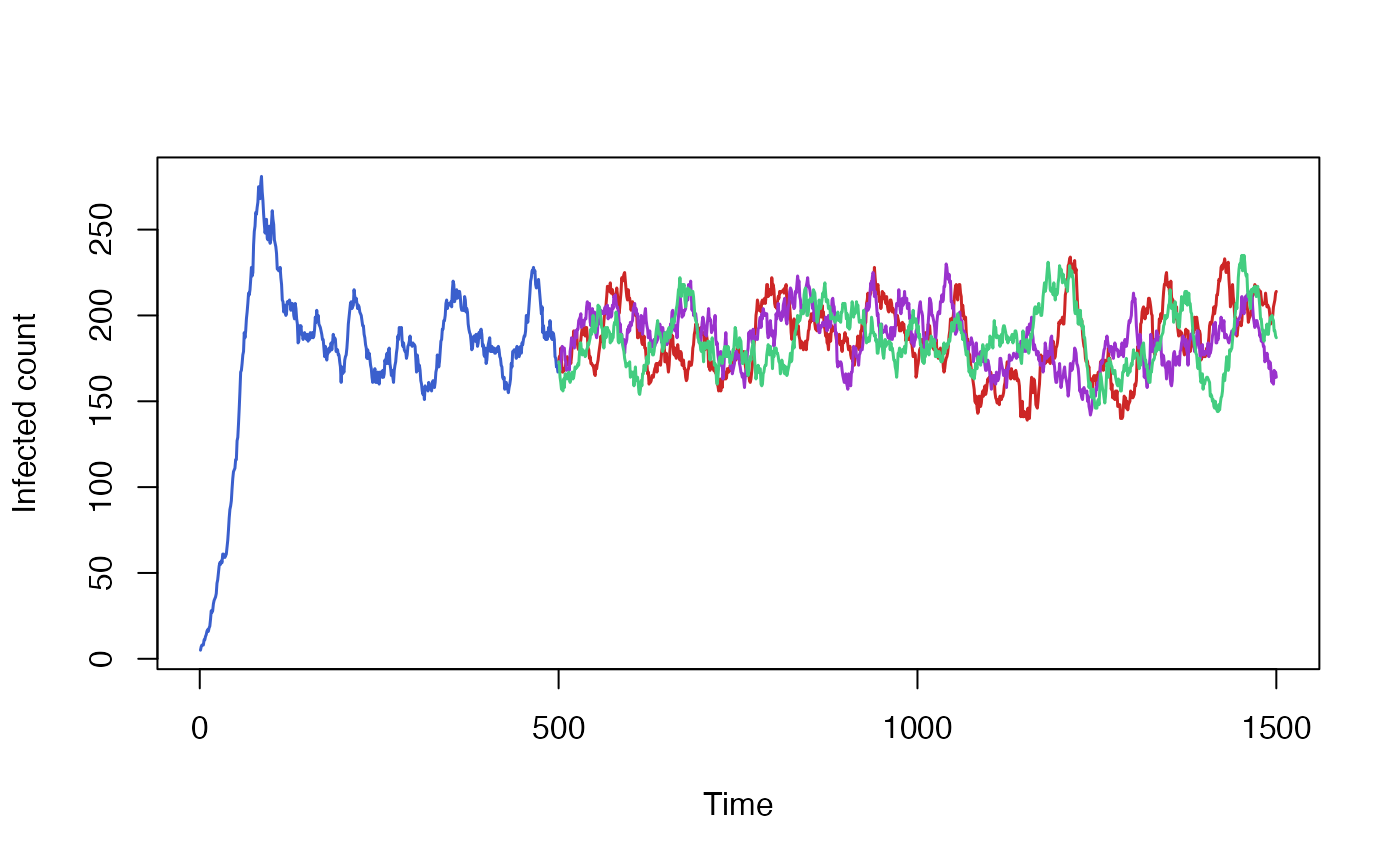
By default resuming a simulation does not restore R’s random number generator’s state. Every resumed run from the same saved state will be independent and, if the model is stochastic, will produce different results.
We can demonstrate that by running the baseline of our SIRS model multiple times and plotting the results. All three runs start off from the same state, inherited from our original model run, but quickly diverge based on the outcome of random draws.
initial <- run_simulation(steps=499)
run1 <- run_simulation(steps = 1500, state = initial$state)
run2 <- run_simulation(steps = 1500, state = initial$state)
run3 <- run_simulation(steps = 1500, state = initial$state)
initial$result[500:1500,] <- NA
matplot(
data.frame(
initial$result[,"I_count"],
run1$result[,"I_count"],
run2$result[,"I_count"],
run3$result[,"I_count"]),
xlab = "Time", ylab = "Infected count",
type = "l", lwd = 1.5, lty = 1, col = colours
)
Sometimes this behaviour may not be desirable, and we would instead like to restore the state of the random number generator exactly where it was when we stopped the first part of the run. One example of this is when checking that our model behaves the same whether or not it was saved and resumed.
The code below show an attempt at running the model twice, once as a continous run and once in a piecewise manner. We would hope that seeding the random generator at the start of the simulation would be enough to get identical results out of it. Unfortunately we don’t, because the random number generator state’s at the intermediate point isn’t being preserved.
set.seed(123)
uninterrupted_run <- run_simulation(steps = 1500)$result
set.seed(123)
piecewise_run_initial <- run_simulation(steps = 499)
piecewise_run_final <- run_simulation(steps = 1500, state = piecewise_run_initial$state)
piecewise_run <- rbind(piecewise_run_initial$result, piecewise_run_final$result[500:1500,])
all.equal(uninterrupted_run, piecewise_run)
#> [1] "Component \"S_count\": Mean relative difference: 0.06844564"
#> [2] "Component \"I_count\": Mean relative difference: 0.1065086"
#> [3] "Component \"R_count\": Mean relative difference: 0.07297747"We can try the same again, but this time set
restore_random_state = TRUE to enable restoring the
simulation state. This time we’ve successfully managed to reproduce the
data from our uninterrupted run.
set.seed(123)
piecewise_run_initial <- run_simulation(steps = 499)
piecewise_run_final <- run_simulation(steps = 1500, state = piecewise_run_initial$state, restore_random_state = TRUE)
piecewise_run <- rbind(piecewise_run_initial$result, piecewise_run_final$result[500:1500,])
all.equal(uninterrupted_run, piecewise_run)
#> [1] TRUEUsing restore_random_state = TRUE resets the global
random number generator’s state, which could have surprising and
undesirable side effects. It is generally useful in tests, but should be
used carefully elsewhere.